
The PCR laboratory

Leukaemia and lymphoma, as with all cancer, is caused by genetic alterations in a number of critical regulatory genes. These abnormalities can be detected directly at a molecular level. Knowledge of these abnormalities can provide prognostic information which can used to guide treatment. The Polymerase Chain Reaction (PCR) provides a rapid means of detecting many of these leukaemia specific aberrations.
Both deoxyribonucleic acid (DNA, the total genetic code present in every cell) and messenger ribonucleic acid (mRNA, the single stranded template used to synthesize protein) can be used as targets for the PCR process.
DNA PCR may be used to demonstrate immunoglobulin heavy chain (IgH) and T-cell receptor (TCR) gene rearrangements. These represent unique clonal markers, and PCR techniques may therefore be used to demonstrate clonality in lymphoproliferative disorders. Additionally, such techniques may be used to detect very low levels of disease; so-called minimal residual disease (MRD) studies.
Chromosomal translocations are common genetic events in leukaemia and lymphoma and result in the dysregulation of a proto-oncogene or alternatively the formation of a chimeric fusion gene. These translocations may be disease defining and often have significant prognostic implications. Many of these translocations are demonstrable by DNA or reverse transcriptase (RT) PCR which uses RNA as a template and can be applied in the routine diagnostic setting as well as in the assessment of MRD.
Clonal IgH rearrangements can be detected by a number of PCR based strategies. These are easy to perform and can be applied to archival BM slides and paraffin embedded tissue specimens as well as DNA derived from fresh PB, BM and tissue.
The ability to demonstrate clonal IgH rearrangements varies between lymphoproliferative disorders and is also dependent on the PCR strategy employed. By end labeling the JH primer with a fluorochrome and analysing the products with an automated fragment analyser equipped with "genescanning" software, definitive clonal rearrangements are routinely detected in 93% of B-lineage ALL and 75% of myeloma cases[1].
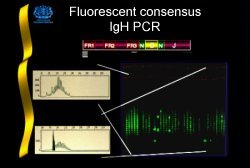
A diagnosis of lymphoid malignancy cannot be made on the basis of a clonal IgH rearrangement alone. Results must be interpreted with caution as clonal rearrangements are demonstrable in a number of inflammatory and infective disorders[2]. Additionally, false negative results may be obtained from diagnostic material derived from patients with well characterised lymphoproliferative disorders. This appears to be a particular problem in multiple myeloma (MM) and non-Hodgkin's lymphoma (NHL) where clonal rearrangements are not demonstrable in approximately 30% of cases[1]. This is likely to be due to the loss of primer binding sites as a result of somatic hypermutation or chromosomal translocation involving the IgH locus.
Minimal residual disease (MRD) refers to the presence of clonal cells in the bone marrow below the level of detection of standard morphological assessment. It has been most widely studied in childhood B-lineage ALL which may therefore be regarded as a "model system" by which different methodologies can be compared.
Assessment of MRD in ALL has traditionally involved sequencing of the IgH rearrangement and the generation of sequence or allele-specific oligonucleotides (ASO) which may be used as probes or PCR primers. These techniques are highly sensitive as they are capable of detecting one leukaemic cell in a background of 100,000 normal cells (0.001%). Despite their sensitivity ASO techniques have a number of disadvantages; they are time consuming, labour intensive and expensive. Additionally, they may not be able to detect clonal change which occurs in up to 30% of cases assessed by Fr3 PCR at both presentation and relapse[3].
In our laboratory we routinely use fluorescent IgH PCR to assess MRD in children with B-lineage ALL, and have sequentially assessed MRD (with fluorescent IgH PCR) in 42 children treated according to the MRC UKALL XI trial protocol and have found that a positive assessment at week 20 of therapy was highly predictive of relapse[4]. These results correlate very well with those obtained by ASOP in a similarly treated group of children with standard risk B-lineage ALL[5].
Fluorescent IgH PCR is a technique ideally suited to routine patient monitoring. The presence of a fluorescent size standard within each lane provides for accurate and reproducible sizing of rearrangements. This significantly aids the identification of clonal cells when they are present within a polyclonal background and also allows for standardisation between laboratories. Results are generally available within one working day which allows for "real time" clinical decision making.
The receptor for antigen on the majority of mature T-cells consists of two polypeptides, termed alpha (a) and beta (b), that are linked by disulphide bonds and are associated with CD3. A small population of mature T-cells express a different TCR heterodimer in association with CD3. This is composed of two polypeptides designated gamma (g) and delta (d) which have a degree of homology with the b and a chains respectively.
We routinely use the TCRg PCR to assess clonality in T-cell disorders. This is the most appropriate single locus to examine; immature T-cell expansions may not have undergone ab recombination whilst the d gene is deleted during a gene recombination.
Clonal rearrangements are commonly visualised by ethidium bromide staining following non denaturing polyacrylamide gel electrophoresis (PAGE). The sensitivity of this technique is of the order of 1%.
Clonal TCRg rearrangements are demonstrable in approximately 70% of patients with T-cell lymphoproliferative disorders. A diagnosis of lymphoid malignancy cannot be made on the basis of a clonal TCRg rearrangement alone however, as false negative results occur in up to 30% of cases while positive assessments can occur in patients with infective or inflammatory disease. Indeed, false positive results appear to occur more commonly with TCRg PCR than with IgH PCR strategies[2,6].
TCRg PCR techniques may be used in the assessment of MRD in patients with ALL. Clonal rearrangements are demonstrable in over 90% of cases of T-lineage ALL but are also demonstrable in approximately 40% of B-lineage cases. T-lineage ALL is rare and consequently TCRg PCR techniques have been more widely applied to the study of B-lineage disease. In this context some groups advocate a multi-locus approach in which each case of B-lineage ALL is assessed with IgH Fr3, TCRg and TCRd PCR techniques[7]. This strategy provides at least one clonal marker in over 90% of cases.
Chromosomal translocations can deregulate genes by two molecular mechanisms; the deregulation of a normal proto-oncogene or the formation of a chimeric fusion gene. Chromosomal translocations which result in the deregulation of a normal proto-oncogene, usually, but not exclusively, involve the genes encoding for antigen receptors (Ig and TCR).
PCR detection of the majority of these aberrations is not possible due to the substantial molecular distances between the oncogene and the antigen receptor gene. An exception however is the detection of the t(14;18) which is demonstrable by DNA-PCR. Demonstration of a t(11;14) in a minority of cases of mantle cell lymphoma (MCL) is also possible using a DNA-PCR strategy[8,9].
| Translocation receptor gene | Leukaemic phenotype | Deregulated oncogene | Antigen |
|---|---|---|---|
| t(8;14)(q24;q32) | Burkitt's NHL | cMYC | IgH |
| t(2:8)(q12;q24) | Burkitt's NHL | cMYC | Ig-kappa |
| t(8;22)(q32;q11) | Burkitt's NHL | cMYC | Igl |
| t(1;14)(q21;q32) | ALL | BCL9 | IgH |
| t(14;18)(q32;q21) | FCLa | BCL2b | IgH |
| t(11:14)(q13;q32) | MCLc | BCL1 | IgH |
| t(14;19)(q32;q13) | B-CLL | BCL3 | IgH |
| t(11;14)(p13;q11) | T-ALL | TCL2d | TCRd |
| t(14;16)(q32;q23) | MMe | c-MAF | IgH |
| t(4;14)(p16;q32) | MM | FGFR3 | IgH |
| t(11;14)(q13;q32) | MM | BCL1?? | IgH |
| t(6;14)(p25;q32) | MM | IRF4 | IgH |
| t(9;14)(p13;q32) | LPLf | PAX5 | IgH |
| inv(14)(q11;q32) | PTCLg | TCRa | IgH |
|
a Follicular centre lymphoma b B-cell leukaemia/lymphoma gene c Mantle cell lymphoma d T-cell leukaemia/lymphoma gene e Multiple Myeloma f Lymphoplasmacytoid Lymphoma g Peripheral T cell lymphoma |
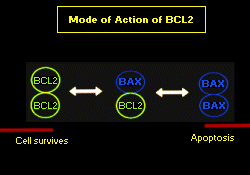
The association of the t(14;18) with Follicle Centre Lymphoma (FCL) was first described cytogenetically by Fukuhara et al[10]. As a consequence of the cloning of a number of t(14;18) bearing cell lines the BCL-2 gene was isolated and subsequently found to inhibit apoptosis.
The translocation involves disruption within the joining region segments on 14q32 and one of two regions within or close to the BCL-2 gene at 18q21. The majority of breakpoints at 18q21 occur within a 150bp segment of the 3' untranslated region of the BCL-2 gene, termed the major breakpoint cluster region (MBR). A second region >20Kb 3' to the MBR is termed the minor cluster region (mcr). Breakpoints within this 500bp segment are found in approximately one third of cases of FCL by Southern analysis.
PCR amplification of this aberration is possible due to the tight clustering of breakpoints at 18q21 and the high degree of sequence homology between the six JH segments at 14q32, allowing a consensus primer to be employed. The PCR strategy we use at HMDS will demonstrate the t(14;18) in approximately 50% of histologically defined FCL. Additional cases may be demonstrated using primers directed at other smaller breakpoint clusters (mcr, 3'MBR and 5'mcr).
PCR detection of the t(14;18) is an attractive proposition for many reasons. Cytogenetics is time consuming, expensive and is frequently unsuccessful, while Southern hybridisation requires relatively large quantities of good quality high molecular weight DNA. A PCR based technique would offer a means of overcoming many of these problems.
At HMDS we routinely use a nested PCR strategy with primers designed for the MBR and JH regions. This strategy demonstrates the t(14;18) in 50-60% of histologically defined FCL. The t(14;18) is also demonstrable in approximately 30% of cases of diffuse large B cell lymphoma (DLBCL) suggesting that these cases may have occurred following transformation of an underlying clinically silent FCL.
Demonstration of the t(14;18) by PCR should not be regarded as diagnostic of lymphoma as it has been demonstrated in normal individuals and those with persistent polyclonal B-cell expansions[11].
The natural history of FCL involves a course of successive relapses with a significant minority of patients transforming to high grade diffuse large B-cell lymphoma, which is unresponsive to treatment. The use of high dose therapy in combination with autologous stem cell rescue has been shown to improve the duration of remission, although no overall improvement in survival has yet been noted. The t(14;18) PCR may be used to assess MRD in those patients that have achieved a complete response following high dose therapy. The significance of molecular residual disease in this setting has yet to be determined.
The t(14;18) PCR may also be used to assess clonal cell contamination of autologous stem cell collections and the purging efficacy of immunological purging strategies. In this regard an inability to purge an autologous bone marrow harvest to PCR negativity appears to result in an increased risk of early relapse.
The majority of chromosomal translocations result in the formation of a fusion gene and consequently a chimeric protein. These proteins show either enhanced activity or a novel function, such as altered DNA binding.
The mechanisms by which these consistent recombinations occur between genes is not fully understood. Although one mechanism may be the recombination of similar repetitive sequences adjacent to the breakpoints on both chromosome partners.
PCR based techniques can be applied to these cases if RNA is extracted and reverse transcribed to a cDNA copy using random hexamers as primers. The strategy at HMDS uses random hexamers as primers for reverse transcription and produces a total cDNA representation from the cells analysed.
Amplification of the normal ABL gene is used as a control to assess the quality of cDNA prior to all RT-PCR procedures. The gene appears to be ubiquitously expressed within all cells and provides a means of determining the RNA quality and/or the success of the reverse transcription step prior to PCR amplification. Amplification of this transcript requires at least 5 x 103 amplifyable Abl molecules to give a positive result when resolved on 2% agarose gel with EtBr staining.
The Philadelphia chromosome (Ph+, 22q-) was the first chromosomal abnormality associated with a specific malignant disease in humans, chronic myeloid leukaemia (CML). We now know that the t(9;22)(q34;q11) results in the formation of two hybrid fusion genes; BCR/ABL on chromosome 22 and ABL/BCR on chromosome 9. The BCR/ABL fusion gene encodes a protein with enhanced tyrosine kinase activity. This process is regarded to be the central mechanism in the pathogenesis of chronic phase CML. The role, if any, of the reciprocal fusion gene ABL/BCR remains uncertain.
The breakpoint in the ABL gene can occur anywhere within a large 300kb segment between exons 1 and 2 at the 5' end of the gene. In the vast majority of CML cases the breakpoint in the BCR gene occurs within a 5.8kb region known as the major breakpoint cluster region (M-bcr). This region spans five exons termed b1-b5 corresponding to exons 12 to 16 of the BCR gene. Regardless of the position of the ABL breakpoint processing results in BCR/ABL mRNA with a b3a2 or b2a2 junction which encode a p210 fusion protein.
In the majority of Ph+ ALL cases the BCR breakpoint occurs further upstream between two alternative exons e2 and e2'. This region is known as the minor breakpoint cluster region (m-bcr). Following splicing an e1a2 junction is formed which encodes a smaller p190 fusion protein.
Rarely the BCR breakpoint occurs between exons e19 and e20; the so-called m-bcr. The resulting e19a2 junction encodes a larger p230 fusion protein.
The t(9;22) is the genetic hallmark of CML. It is demonstrable by conventional cytogenetics in 90% of cases while a further 5% will have variant translocations, both cases the molecular consequence of the translocation is the formation of a BCR/ABL fusion gene.
BCR/ABL fusion transcripts are also demonstrable in a significant minority of patients with ALL. Fusion transcripts are significantly commoner in adults (approximately 30%) than children (approximately 5%) and their presence confers a very poor prognosis. In approximately 2/3 cases a p190 associated transcript is generated and may signify true de novo ALL. The remaining cases have p210 associated transcripts suggesting that in these cases the disease occurred as a result of blastic transformation of "clinically silent" chronic phase CML.
RT-PCR techniques have been extensively used to assess MRD in CML patients following allogeneic transplantation. BCR/ABL transcripts are detectable in many patients for up to 9 months following transplant but their presence does not appear to predict relapse.
Quantitation of BCR/ABL transcripts in this instance is now possible using either a plasmid competitor titration assay, or more recently at HMDS using "real time" quantitation and Taqman technology.
Increasing levels of BCR/ABL transcripts appears to predict relapse in this setting and the titration assay is commonly used to guide therapy with donor lymphocyte infusions. MRD may also be assessed in patients who have achieved a complete cytogenetic response following a-interferon therapy.
RT-PCR techniques may also be applied to patients with Ph+ ALL following conventional and high-dose therapy. There appears to be a significant practical difference between CML and Ph+ ALL. In the former the level of residual disease appears to be very similar in the bone marrow and peripheral blood allowing regular patient monitoring without the need for invasive marrow sampling. In Ph+ ALL the bone marrow appears to be significantly more informative and is the preferred tissue of study.
The Medical Research Council AML X trial demonstrated superior survival in patients presenting with either a t(15;17), t(8;21) or inv(16). This data has been used to modify treatment in the current AML XII trial such that patients demonstrating these abnormalities are no longer selected for the transplant arm of the trial.
Acute Promyelocytic Leukaemia (APL; AML M3) is identified by the unique t(15;17) translocation which fuses the PML gene to the retinoic acid receptor a gene (RARA). The translocation is exclusively associated with APL and confers a good prognosis.
The inversion of chromosome 16 between 16q22 and 16p13 occurs primarily in cases of AML M4 with associated eosinophilia. Inversion fuses the smooth muscle myosin heavy chain gene (MYH11) to the core binding factor beta gene (CBFb), a T cell transcription factor.
The t(8;21) is associated primarily with AML M2 but is also seen in other FAB types and MDS. The translocation fuses the AML1 gene at 21q22 with the ETO gene at 8q22.
Both fusion gene products associated with APL, the PML/RARa from the derivative chromosome 15 and RARa/PML from the derivative chromosome 17 can be demonstrated by RT-PCR. Breakpoints within RARa are generally conserved but alternative breakpoints occur within the PML gene. Disruption within intron 3 of PML (bcr3) are termed 5' breakpoints, whereas 3' breakpoints occur within intron 6 (bcr1) or less commonly in the region between exons 5 and 6 (bcr2). 3' breakpoints (bcr1 and bcr2) generate different sized products when amplified using nested primers to exons 3 of both PML and RARa. This occurs as a consequence of the alternate splicing of PML exons immediately 5' to the breakpoint. Interpretation of gel patterns can be difficult in this situation. However, once a 3' breakpoint is established the second round amplification can be modified by replacing the PML exon 3 primer with a primer designed to exon 5. This strategy makes gel interpretation easier as a single product will be demonstrated when breakpoints occur within the bcr1 and bcr2 regions whereas no product will be generated in cases with 5' breakpoints.
RT-PCR detection of the reciprocal RARa/PML fusion product is possible in approximately 75% of APL cases. The detection of this second fusion product is desirable as it provides useful confirmatory information.
We amplify the 5'CBFb - 3' MYH11 message rather than the derivative 5' MYH11 - 3'CBFb transcript. The hybrid gene contains all but 17 codons of the b gene fused to the MYH11 gene. Breakpoints are conserved within the b gene and have been reported at three sites within the MYH11 gene. This results in four possible chimeric transcripts, (A-D). A transcripts however account for over 90% of cases and therefore a single set of nested primers is used to span this region.
We amplify the AML1-ETO(MTG8) fusion gene from the derivative chromosome 8. Breakpoints are clustered within a single intron of the AML1 gene immediately 3' to the runt homology domain. Nested primers were designed from published cDNA sequence data. Breakpoints within the ETO gene are also clustered and nested primers were designed to the ETO common exon. Alternative splicing of ETO exons immediately 3' to the breakpoint have been reported which result in the amplification of a number of larger fragments.
As the identification of these abnormalities is used to alter therapy, a robust procedure capable of detecting all positive cases at presentation is essential. RT-PCR offers advantages over conventional cytogenetics which may underestimate the true incidence of these translocations due to various biological and technical problems:
With the exception of the t(15;17) in APL these aberrations are not FAB specific. It is therefore critical to analyse all cases of AML at presentation regardless of their morphological appearance.
Rapid detection of the t(15;17) is essential as it defines a group of patients responsive to all-trans retinoic acid (ATRA) differentiation therapy. Cases of APL lacking the t(15;17) or with variant translocations appear to respond poorly to this treatment.
Approximately 70% of APL cases show 3' PML breakpoints and it has been reported that these patients have an improved prognosis with an improved DFS compared to those with 5' breakpoints. Discrimination between the bcr1 and bcr2 breakpoints may also have important prognostic implications as cases presenting with bcr2 breakpoints appear to have reduced in-vitro sensitivity to ATRA.
The reciprocal RARa/PML fusion product is expressed in 67% to 81% of APL cases. Detection of these transcripts by RT-PCR is possible and provides important confirmation of the diagnosis.
RT-PCR may be used to monitor MRD in AML patients following conventional and high dose therapy. MRD may be demonstrated in many patients in long term clinical remission. AML1/ETO, PML/ RARa and CBFb/MYHII transcripts have all been demonstrated in long term remitters. There is therefore a great deal of controversy as to whether PCR negativity should be a treatment goal, and this would also suggest that qualitative PCR is of limited clinical value.
Quantitation of the levels of AML1/ETO, PML/ RARa and CBFb/MYHII have recently been described by our group and others, in patients presenting with an t(8;21), t(15;17) and inv(16). Further advances in the quantitation of these chimeric fusion genes have recently been achieved by utilising "real time" quantitation with the Taqman assay combined with the ABI 7700 platform.
Risk adapted therapy is also being applied to patients in large scale randomised clinical trials in ALL. Demonstration at presentation of a number of clinical and biological risk factors allow patients to be divided into different risk groups. Age and presenting white cell count are still the most significant predictors of outcome in this disease. However, identification of a number of common genetic lesions can also provide independent prognostic information. Demonstration of a t(9;22) at presentation is associated with a very poor prognosis in both children and adults with this disease. Allogeneic transplantation is now considered the most appropriate first line treatment for this group of patients.
The t(12;21)(p13;q22) has recently been described in ALL. This results in the formation of a fusion gene with the combination of the AML1/CBFa gene and TEL gene, an ETS like putative transcription factor. The translocation appears cryptic to normal karyotyping techniques but has been demonstrated in 16-26% of childhood ALL by RT-PCR making it by far the commonest translocation associated with this disease. The translocation is almost exclusively found in patients with the common ALL (c-ALL) immunophenotype presenting between the ages of 1-10. It appears to confer an excellent prognosis.
We use a nested strategy to demonstrate the t(12;21) in cases of ALL. Nested primers are directed against exons 5 and 3 of TEL and AML1 respectively. Chimeric TEL/AML1 transcripts show a positive band of 267bp by this nested procedure. An additional 228bp fragment is frequently seen and occurs as a result of the removal of exon 2 of the AML1 gene by alternative splicing. This smaller fragment may occur rarely in isolation as a result of fusion of TEL exon 5 with exon 3 of AML1.
The t(12;21) is demonstrable in less than 0.1% of cases by routine cytogenetics but in up to 26% by RT-PCR. The abnormality is cryptic to conventional cytogenetics due to the banding pattern at the breakpoint regions on both chromosomes 12 and 21. The abnormality appears to be associated with a cALL phenotype and confers a good prognosis. Identification of this translocation is clearly important and the RT-PCR provides the most appropriate technique.
A small number of studies have used the t(12;21) RT-PCR to monitor MRD in children with ALL. Two studies have independently demonstrated the chemo-sensitive nature of this leukaemia, with the majority of patients in both studies reverting to PCR negativity following induction therapy. More appropriate clinical information may be gained from quantitative studies. Indeed, Nakao et al, have recently used a competitor titration assay to assess the level of residual disease in two patients with a t(12;21).
Comments & feedback to: [email protected]